Note
Go to the end to download the full example code.
Download audio files from Xeno-Canto and automatically extract characteristics
The goal of this example is to show how to automatically download audio files from Xeno-Canto and process them to automatically extract audio characteristics in order to classify the sound made by different species. We focus on the characteristics of the drumming performed by the woodpeckers species that are present in Europe.
Dependencies: To execute this example you need to have installed the librosa, sklearn and pandas Python packages.
(from https://woodpeckersofeurope.blogspot.com/2007/11/drumming.html) Woodpeckers of Europe 10 species of woodpecker (Picidae) breed in Europe: 9 resident species and the migratory Wryneck. 8 of these 10 also occur outside Europe, with the distribution of Eurasian Three-toed, White-backed, Lesser Spotted, Great Spotted, Black & Grey-headed Woodpeckers stretching eastwards from the Western Palearctic into Asia, whilst Syrian is found in the Middle East & Asia Minor & Wryneck winters in Africa. The global ranges of Green & Middle Spotted Woodpeckers are confined to the Western Palearctic.
Eurasian Three-toed : Picoides tridactylus White-backed : Dendrocopos leucotos Lesser Spotted : Dryobates minor Great Spotted : Dendrocopos major Black : Dryocopus martius Grey-headed : Picus canus Syrian : Dendrocopos syriacus Wryneck : Jynx torquilla Green : Picus viridis Middle Spotted : Dendrocoptes medius
# sphinx_gallery_thumbnail_path = './_images/sphx_glr_plot_woodpecker_drumming_characteristics_002.png'
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib.lines import Line2D
import numpy as np
from pathlib import Path
import sys
import os
import time
import warnings
# suppress all warnings
warnings.filterwarnings("ignore")
from scipy import signal
import librosa
import librosa.display
from sklearn.manifold import TSNE
from sklearn.preprocessing import StandardScaler
from maad import sound, util, rois
Define local function
function to grab all audio files in a folder
Define constants
Directory where to download the audiofile from xeno-canto
XC_ROOTDIR = '../../data/'
Name of the dataset. This will be used to create a subdir with the same name
XC_DIR = 'woodpecker_dataset'
data = [['Eurasian Three-toed', 'Picoides tridactylus'],
['White-backed', 'Dendrocopos leucotos'],
['Lesser Spotted', 'Dryobates minor'],
['Great Spotted', 'Dendrocopos major'],
['Black', 'Dryocopus martius'],
['Grey-headed', 'Picus canus'],
['Syrian', 'Dendrocopos syriacus'],
['Wryneck', 'Jynx torquilla'],
['Green', 'Picus viridis'],
['Middle Spotted', 'Dendrocoptes medius']]
Query Xeno-Canto
get the genus and species needed for Xeno-Canto
Build the query dataframe with columns paramXXX gen : genus cnt : country area : continent (europe, america, asia, africa) q : quality len : length of the audio file type : type of sound : ‘song’ or ‘call’ or ‘drumming’ Please have a look here to know all the parameters and how to use them : https://xeno-canto.org/help/search
df_query = pd.DataFrame()
df_query['param1'] = gen
df_query['param2'] = sp
df_query['param3'] ='type:drumming'
df_query['param4'] ='area:europe'
# df_query['param5 ='len:"5-120"'
# df_query['param6'] ='q:">C"'
# Get recordings metadata corresponding to the query
df_dataset= util.xc_multi_query(df_query,
format_time = False,
format_date = False,
verbose=True)
Loading page 1...
https://www.xeno-canto.org/api/2/recordings?query=Picoides%20tridactylus%20type:drumming%20area:europe&page=1
Found 1 pages in total.
Saved metadata for 251 files
Loading page 1...
https://www.xeno-canto.org/api/2/recordings?query=Dendrocopos%20leucotos%20type:drumming%20area:europe&page=1
Found 1 pages in total.
Saved metadata for 193 files
Loading page 1...
https://www.xeno-canto.org/api/2/recordings?query=Dryobates%20minor%20type:drumming%20area:europe&page=1
Loading page 2...
https://www.xeno-canto.org/api/2/recordings?query=Dryobates%20minor%20type:drumming%20area:europe&page=2
Found 2 pages in total.
Saved metadata for 532 files
Loading page 1...
https://www.xeno-canto.org/api/2/recordings?query=Dendrocopos%20major%20type:drumming%20area:europe&page=1
Loading page 2...
https://www.xeno-canto.org/api/2/recordings?query=Dendrocopos%20major%20type:drumming%20area:europe&page=2
Found 2 pages in total.
Saved metadata for 957 files
Loading page 1...
https://www.xeno-canto.org/api/2/recordings?query=Dryocopus%20martius%20type:drumming%20area:europe&page=1
Found 1 pages in total.
Saved metadata for 353 files
Loading page 1...
https://www.xeno-canto.org/api/2/recordings?query=Picus%20canus%20type:drumming%20area:europe&page=1
Found 1 pages in total.
Saved metadata for 105 files
Loading page 1...
https://www.xeno-canto.org/api/2/recordings?query=Dendrocopos%20syriacus%20type:drumming%20area:europe&page=1
Found 1 pages in total.
Saved metadata for 33 files
Loading page 1...
https://www.xeno-canto.org/api/2/recordings?query=Jynx%20torquilla%20type:drumming%20area:europe&page=1
Found 1 pages in total.
Saved metadata for 0 files
Loading page 1...
https://www.xeno-canto.org/api/2/recordings?query=Picus%20viridis%20type:drumming%20area:europe&page=1
Found 1 pages in total.
Saved metadata for 36 files
Loading page 1...
https://www.xeno-canto.org/api/2/recordings?query=Dendrocoptes%20medius%20type:drumming%20area:europe&page=1
Found 1 pages in total.
Saved metadata for 26 files
Download audio Xeno-Canto
From the metadata that was collected in the previous section, select a maximum of 20 files per species, regarding the quality and the length
df_dataset = util.xc_selection(df_dataset,
max_nb_files=20,
max_length='01:00',
min_length='00:10',
min_quality='B',
verbose = True )
Picoides tridactylus
... request 20 files of quality A
--> found 20 files of quality A and 00:10<length<01:00
total files : 20
-----------------------------------------
Dendrocopos leucotos
... request 20 files of quality A
--> found 13 files of quality A and 00:10<length<01:00
... request 7 files of quality B
--> found 7 files of quality B and 00:10<length<01:00
total files : 20
-----------------------------------------
Dryobates minor
... request 20 files of quality A
--> found 20 files of quality A and 00:10<length<01:00
total files : 20
-----------------------------------------
Dendrocopos major
... request 20 files of quality A
--> found 20 files of quality A and 00:10<length<01:00
total files : 20
-----------------------------------------
Dryocopus martius
... request 20 files of quality A
--> found 15 files of quality A and 00:10<length<01:00
... request 5 files of quality B
--> found 5 files of quality B and 00:10<length<01:00
total files : 20
-----------------------------------------
Picus canus
... request 20 files of quality A
--> found 19 files of quality A and 00:10<length<01:00
... request 1 files of quality B
--> found 1 files of quality B and 00:10<length<01:00
total files : 20
-----------------------------------------
Dendrocopos syriacus
... request 20 files of quality A
--> found 11 files of quality A and 00:10<length<01:00
... request 9 files of quality B
--> found 5 files of quality B and 00:10<length<01:00
total files : 16
-----------------------------------------
Picus viridis
... request 20 files of quality A
--> found 0 files of quality A and 00:10<length<01:00
... request 20 files of quality B
--> found 7 files of quality B and 00:10<length<01:00
total files : 7
-----------------------------------------
Dendrocoptes medius
... request 20 files of quality A
--> found 5 files of quality A and 00:10<length<01:00
... request 15 files of quality B
--> found 5 files of quality B and 00:10<length<01:00
total files : 10
-----------------------------------------
download all the audio files into a directory with a subdirectory for each species
util.xc_download(df_dataset,
rootdir = XC_ROOTDIR,
dataset_name= XC_DIR,
overwrite=False,
save_csv= True,
verbose = True)
***WARNING*** : The directory ../../data already exists
Grab all audio filenames in the directory
create a dataframe with all recordings in the directory
filelist = grab_audio(XC_ROOTDIR+XC_DIR)
Create new columns with short filename and species names
df = pd.DataFrame()
for file in filelist:
df = pd.concat([df, pd.DataFrame({
'fullfilename': [file],
'filename': Path(file).parts[-1][:-4],
'species': Path(file).parts[-2]
})
],
ignore_index=True)
print('=====================================================')
print('number of files : %2.0f' % len(df))
print('number of species : %2.0f' % len(df.species.unique()))
print('=====================================================')
=====================================================
number of files : 26
number of species : 2
=====================================================
Process all audio files, species by species
In this part, all audio file will be processed in order to extract each drumming portion separately. Then pulses are automaticaly detected for each drumming before computing drumming parameters such as median pulse rate, duration, number of pulses…
# store starting time
start_time = time.time()
# Create an empty dataframe to drummings parameters
df_drums = pd.DataFrame()
# Loop to extract portion of the dataframe corresponding to a single species
for species in df.species.unique():
# get the dataframe corresponding to the current species
current_df = df[df.species == species]
# Display the current species
print ('\n')
print (' %s ' %species)
# Loop to load and process each audio file of the current species
fullfilename_list = list(current_df.fullfilename)
idx = 0
for fullfilename in fullfilename_list:
# Create a temporary dataframe in order to store the parameters of
# the drummings found in the current audio file
df_drums_temp = pd.DataFrame()
# extract audio filename and species
path, filename_with_ext = os.path.split(fullfilename)
_, species = os.path.split(path)
file = os.path.splitext(filename_with_ext)[0]
# 1. load audio
s, fs = librosa.load(fullfilename, sr=16000)
# 2. save parameters in dataframe for each file
df.loc[df.fullfilename == fullfilename, 'length'] = len(s)
df.loc[df.fullfilename == fullfilename, 'sampling_freq'] = fs
# 3. bandpass filter around drumming frequencies
fcut = (25, 5000)
s = sound.select_bandwidth(s,
fs,
fcut=fcut,
forder=1,
fname='butter',
ftype='bandpass')
# 4. find portions of the signal (ROIs) that contain a drumming
df_rois = rois.find_rois_cwt(s,
fs,
flims=(50,4000),
tlen=4,
th=1e-3,
display=False)
# 5. Loop to process each ROI previously found
pulseRateMedian = []
drum_duration = []
n_pulses = []
interval_min = []
interval_max = []
interv1_intervMax = []
intervL_intervMax = []
MeanAcc = []
Amp_pulses_min = []
AMp_pulses_first = []
Amp_pulses_last = []
# Loop
for index, row in df_rois.iterrows():
# trim sound to process only portion of the sound that correspond
# to the current ROI
s_trim = sound.trim(s, fs, row['min_t'], row['max_t'])
s_trim = s_trim - np.mean(s_trim)
s_trim = s_trim / np.max(np.abs(s_trim))
# compute fast enveloppe with windows of 32 samples
env = sound.envelope(s_trim, Nt=32)
if np.median(env) < 0.1 :
pulses, pulses_info = signal.find_peaks(env, distance=15, height = np.median(env)*2, prominence=0.2)
# convert pulses in sample into seconds
pulses = pulses/fs*32
# get the relative pulse amplitude
pulse_heights = pulses_info['peak_heights']
if (len(pulses) > 10) and (np.max(np.diff(pulses))<0.2) and (1/np.median(np.diff(pulses))>10):
pulseRateMedian += [1/np.median(np.diff(pulses))]
drum_duration += [pulses[-1] - pulses[0]]
n_pulses += [len(pulses)]
interval_min += [np.min(np.diff(pulses))]
interval_max += [np.max(np.diff(pulses))]
interv1_intervMax += [pulses[1] - pulses[0]]
intervL_intervMax += [pulses[-2] - pulses[-1]]
MeanAcc += [np.mean(np.diff(np.diff(pulses)))]
Amp_pulses_min += [np.min(pulse_heights)]
AMp_pulses_first += [pulse_heights[0]]
Amp_pulses_last += [pulse_heights[-1]]
# plot some envelopes with peak detection
# if idx%10 == 0 :
# plt.figure()
# plt.plot(env)
# plt.plot(pulses*fs/32, env[(pulses*fs/32).astype('int')], "x")
# plt.show()
else :
pulseRateMedian += [np.nan]
drum_duration += [np.nan]
n_pulses += [np.nan]
interval_min += [np.nan]
interval_max += [np.nan]
interv1_intervMax += [np.nan]
intervL_intervMax += [np.nan]
MeanAcc += [np.nan]
Amp_pulses_min += [np.nan]
AMp_pulses_first += [np.nan]
Amp_pulses_last += [np.nan]
if len(df_drums) == 0 :
df_drums['pulseRateMedian'] = pulseRateMedian
df_drums['drum_duration'] = drum_duration
df_drums['n_pulses'] = n_pulses
df_drums['interval_min'] = interval_min
df_drums['interval_max'] = interval_max
df_drums['interv1_intervMax'] = interv1_intervMax
df_drums['intervL_intervMax'] = intervL_intervMax
df_drums['MeanAcc'] = MeanAcc
df_drums['Amp_pulses_min'] = Amp_pulses_min
df_drums['AMp_pulses_first'] = AMp_pulses_first
df_drums['Amp_pulses_last'] = Amp_pulses_last
df_drums['species'] = species
df_drums['filename'] = file
else:
df_drums_temp['pulseRateMedian'] = pulseRateMedian
df_drums_temp['drum_duration'] = drum_duration
df_drums_temp['n_pulses'] = n_pulses
df_drums_temp['interval_min'] = interval_min
df_drums_temp['interval_max'] = interval_max
df_drums_temp['interv1_intervMax'] = interv1_intervMax
df_drums_temp['intervL_intervMax'] = intervL_intervMax
df_drums_temp['MeanAcc'] = MeanAcc
df_drums_temp['Amp_pulses_min'] = Amp_pulses_min
df_drums_temp['AMp_pulses_first'] = AMp_pulses_first
df_drums_temp['Amp_pulses_last'] = Amp_pulses_last
df_drums_temp['species'] = species
df_drums_temp['filename'] = file
df_drums = pd.concat([df_drums,df_drums_temp],
ignore_index=True)
# counter
sys.stdout.write('\r')
sys.stdout.write('%2.0f%%' %np.round(((idx+1)/len(current_df)*100)))
idx = idx+1
print("--- %2.2f minutes ---" % ((time.time() - start_time)/60))
# drop all rows with NaN
df_drums = df_drums.dropna()
Picoides tridactylus_Eurasian Three-toed Woodpecker
5%
10%Warning: No detection found
15%Warning: No detection found
20%
25%
30%
35%
40%Warning: No detection found
45%
50%
55%Warning: No detection found
60%
65%
70%
75%
80%
85%Warning: No detection found
90%
95%
100%
Dendrocopos leucotos_White-backed Woodpecker
17%
33%Warning: No detection found
50%Warning: No detection found
67%Warning: No detection found
83%
100%--- 0.09 minutes ---
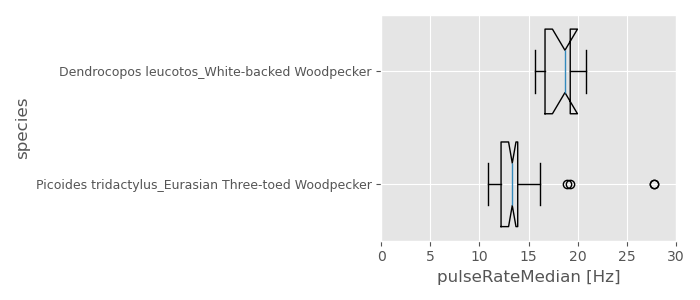
Display boxplot
Display a boxplot of the feature “pulseRateMedian” for each species
plt.style.use('ggplot')
# create a figure
fig = plt.figure(figsize= (7,3))
ax = fig.add_subplot(111)
n = 0
# loop to build a boxplot for each species based on the feature "pulseRateMedian"
for species in df_drums.species.unique():
ax.boxplot(df_drums[df_drums.species == species]['pulseRateMedian'],
positions=[n+1],
widths = 0.75,
vert = False,
notch=True)
n += 1
ax.set_yticks(np.arange(1,len(df_drums.species.unique())+1))
ax.set_yticklabels(df_drums.species.unique(),
fontsize=9)
ax.set_xlabel('pulseRateMedian [Hz]')
ax.set_ylabel('species')
ax.set_xlim(0,30)
plt.tight_layout()
plt.show()
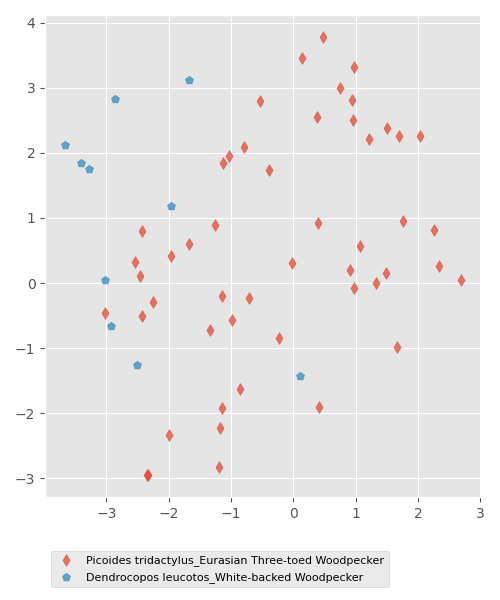
Display clusters based on the drummings features
A collection of features is associated to each drumming found in the audio recordings. The goal is to display clusters in 2D with the dimensionality reduction tool t-SNE and associate a color to each point that corresponds to the belonging species. It is then possible to observe species that are clearly grouped into separate clusters from species that are mixed with others
# Preprocess data : data scaler
df_drums = df_drums.dropna()
X = df_drums.drop(columns=['species','filename'])
scaler = StandardScaler()
X = scaler.fit_transform(X)
# compute the dimensionality reduction
tsne = TSNE(n_components=2,
perplexity=30,
init='pca',
n_iter = 5000,
n_jobs = -1,
verbose=True)
Y = tsne.fit_transform(X)
# overlay all species
plt.figure(figsize=(5,6))
g = []
markers = Line2D.filled_markers
for species in df_drums.species.unique():
g.append(plt.scatter(Y[(df_drums['species'] == species), 0],
Y[(df_drums['species'] == species), 1],
marker = markers[np.random.randint(0, len(markers))],
alpha=0.75))
plt.legend(g,
df_drums.species.unique(),
bbox_to_anchor=(0, -0.1),
loc='upper left',
fontsize=8,
frameon = True)
plt.tight_layout()
[t-SNE] Computing 59 nearest neighbors...
[t-SNE] Indexed 60 samples in 0.000s...
[t-SNE] Computed neighbors for 60 samples in 0.001s...
[t-SNE] Computed conditional probabilities for sample 60 / 60
[t-SNE] Mean sigma: 2.428003
[t-SNE] KL divergence after 250 iterations with early exaggeration: 46.562538
[t-SNE] KL divergence after 1200 iterations: 0.114990
Total running time of the script: (1 minutes 24.466 seconds)